About the service
Detailed information regarding the BoneDVC service:
Accurate bone digital volume correlation service
Scientific background
The measurement of bone deformation under loading is important to understand the effect of musculoskeletal pathologies and to optimise their treatments.
The Digital Volume Correlation (DVC) approach applied to the bone tissue, proposed first by Bay et al. (1999) is the only method to provide a measurement of the full-field 3D deformation. It combines an elastic registration algorithm to compute the displacement to transform the undeformed image into the deformed one, and the differentiation of the displacement fields into strains. The main challenge in applying this method to bone tissue is that bone fails at small deformations (approximately 1% as reported by Bayraktar et al., 2004). Therefore, the expected strains in the elastic range are small and the DVC method needs to be very accurate.
The BoneDVC service uses the algorithm proposed by Dall'Ara et al. (2014), which has been tested for several bone structures and was found to be more accurate than other published or commercially available alternative (Dall'Ara et al., 2017; Palanca et al., 2015). The user is encouraged to look into the above-mentioned publications for the details of the algorithm. The accuracy of the BoneDVC algorithm has been tested on several bone structures images with standard desktop microCT (Palanca et al., 2016; Tozzi et al., 2017; Giorgi & Dall'Ara, 2018) or high resolution Synchrotron microCT (Palanca et al., 2017; Comini et al., 2019).
This algorithm has been also used for validating the local predictions of finite element models for different bone microstructures (Kusins et al., 2019; Oliviero et al., 2018; Costa et al., 2017; Chen et al., 2017).
BoneDVC service
The BoneDVC service estimates the field of displacements and strains in bone specimens, provided two 3D images of the undeformed and deformed bones (Figure 1).
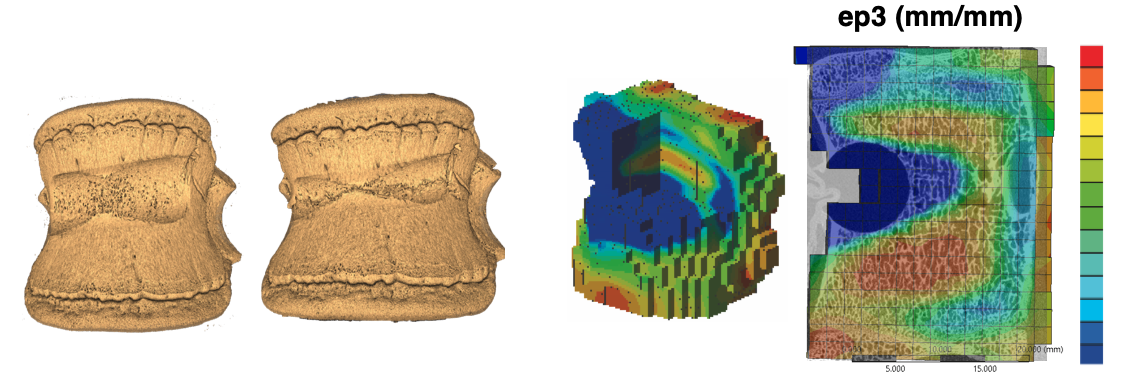
Figure 1: example of application of the BoneDVC service to study the deformation of vertebral bodies with induced lesions (courtesy of Dr Marco Palanca).
A user uploads two 3D images, usually micro-Computed Tomography (microCT) scans of the specimen that has been tested in situ within the scanning machine. The first image represents the undeformed configuration ("Fixed_Image"), the second represents the deformed configuration ("Moved_Image"). As option, the user can also upload a mask ("Mask_Image") that specifies where the registration should take place. The threshold in the mask image ("Thresh") represents the limit in the grey-values above which each voxel is considered included in the region of interest where the registration should be run.
Images should be uploaded as 8-bit multi-slice DICOM files (1 file for each 3D stack) and should have the same size (voxels in every Cartesian direction). The images should have been already rigidly co-registered and the results will be provided with respect to the coordinate system of the input images. At the moment the limit of the input images is approximately 2Gb each, but this will be increased in the future with a parallel version of the elastic registration algorithm currently under development.
A DVC job is created by clicking on New Job and submitting a DVC configuration file (Figure 3; example below). This will generate a form in which a VOI's Fixed, Moved and Mask Dicom images can be selected for the corresponding Specimen (Figure 2A). Files can be selected either from Google Drive* or your Desktop. The results will be provided in the structure as reported in Figure 2B. If required, multiple specimens and VOIs can be specified in a single configuration file.
Google Drive folder structures
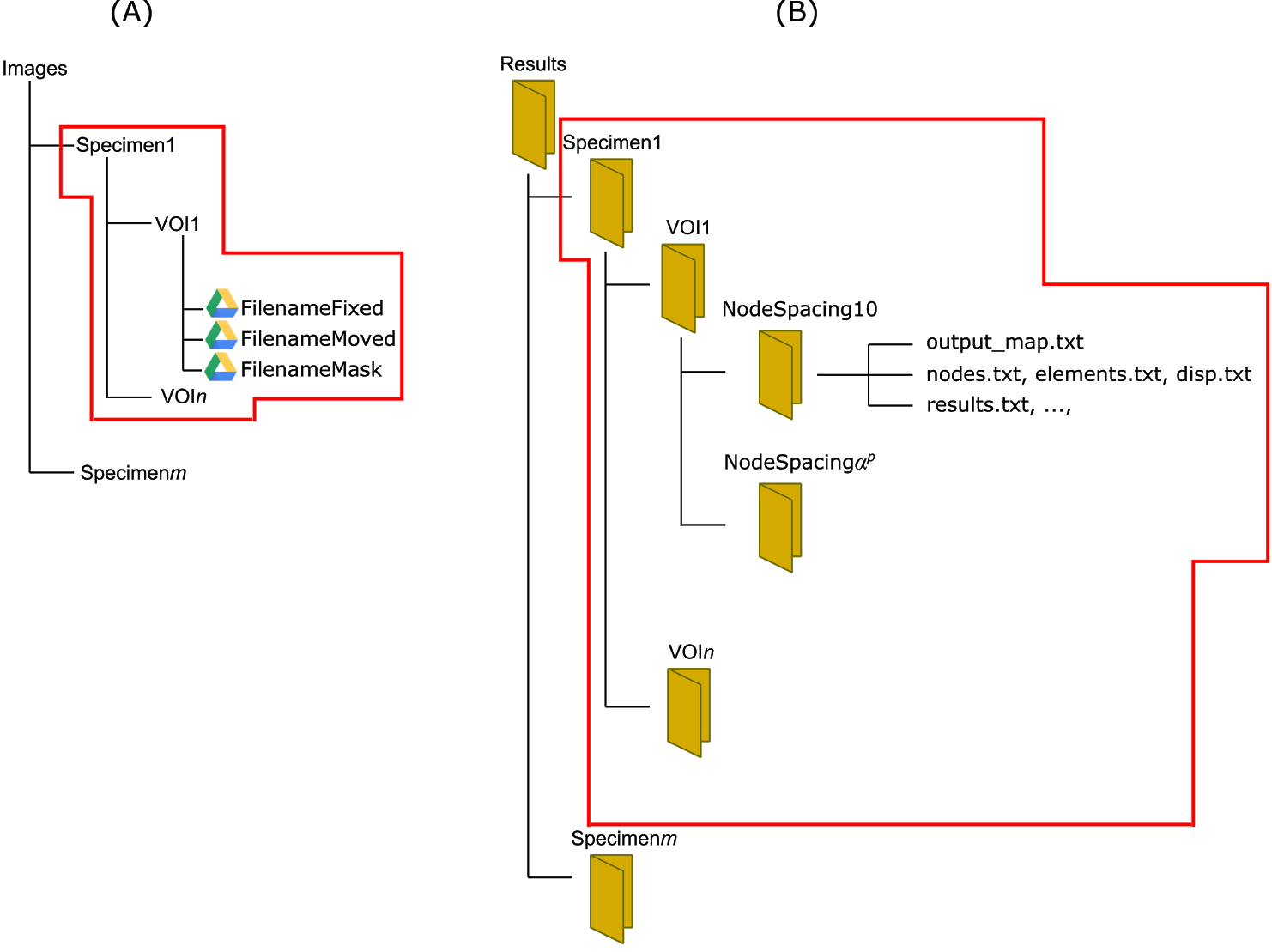
Figure 2: structure of the folders for input images and results.
* A Sheffield University Google account is required to access this service. When you use the Google Drive file picker for the first time this website will ask your permission to add the BoneDVC app to your Google Drive account. You can remove this app at any time in Google Drive by going to:
Settings > Managing Apps > Options > Disconnect from Drive
If you do not have a Sheffield University Google account please contact this email address to access this service online.
The user needs to provide a configuration file to specify the input parameters and the registrations. A typical configuration file looks like the one below and can be downloaded here.
Example configuration file

Figure 3: Typical example of configuration file. A template for the configuration file can be downloaded in the service.
Description of required configuration file entries
| Project | name of the considered project |
| Vox_Dim | dimension of the voxel in the images to be registered in microns |
| Specimen_Spec1 | name of the first specimen to be registered (more specimens can be provided) |
| VOIs_Spec1 | name of the volume of interest (VOI) to be analysed within the first specimen (more VOIs can be provided for the same specimen) |
| Thresh_Spec1 | threshold value for the identification of the region to be registered in the mask file |
| NSs_Spec1 | values of the nodal spacing (NS, in number of voxels) for which the registrations will be run (more than one value can be provided) |
| Iter_Spec1 | number of iterations after which to stop the registration algorithm in case it has not converged (different values for different NSs can be provided). Default value is 100. |
| Results | code for deciding whether to run the registration with (1) or without (0) mask image |
The algorithm will automatically compute the field of displacements by using deformable registration and the strain after differentiation of the displacement field. The user will receive the outputs in the project folder according to the chosen Specimens, VOIs and NSs.
Description of results files
| disp.txt | list of displacements applied to each node (from output_map.txt) |
| elements.txt | list of elements |
| nodes.txt | list of node coordinates |
| output_map.txt | list of nodes coordinates and corresponding displacements along x, y and z directions |
| results_elements.txt | list of principal strains computed at the element centroids. The first column contains the list of element numbers, while the next columns contain the list of first, second and third principal strains respectively |
| results_elements_xyz.txt | list of strains in the three directions computed at the element centroids |
| results.txt | list of principal strains computed at the nodes. The first column contains the list of node numbers, while the next columns contain the list of first, second and third principal strains respectively |
| results_xyz.txt | list of strains in the three directions computed at the nodes |
| configuration_file.txt | configuration file used for the analyses |
| cronLog-*.txt | log file(s) written during the analyses |